BrainDrugs consists of seven coherent work packages, as depicted in the figure below.
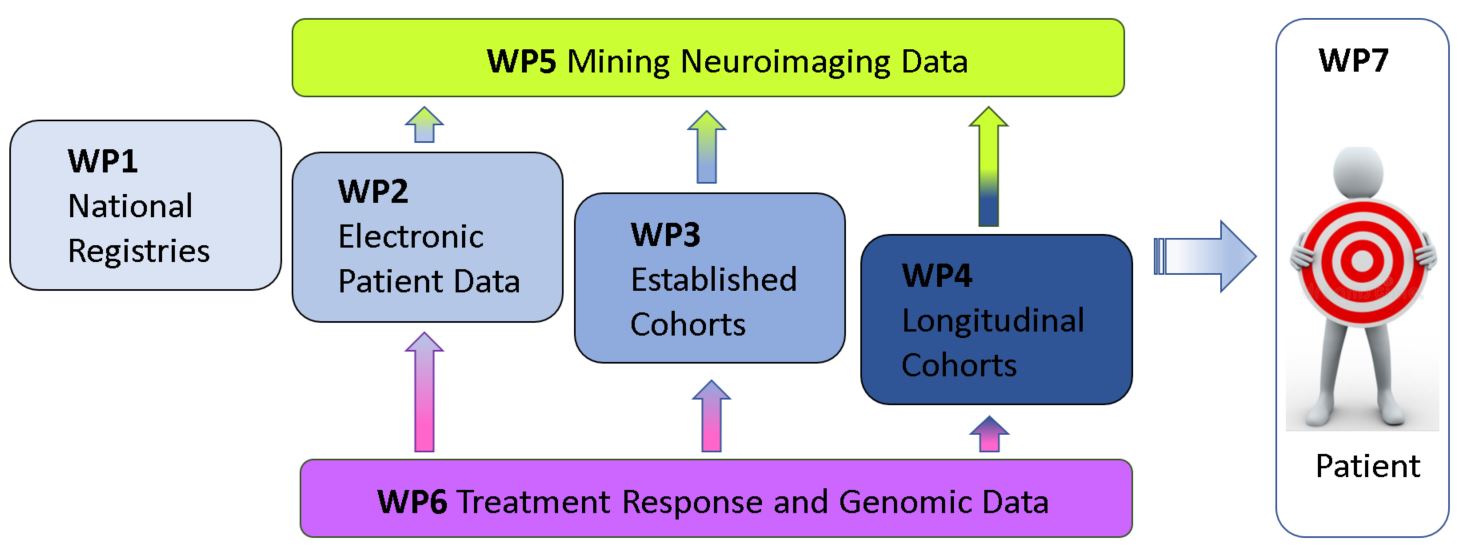
WP1. National Registries
WP-leader: Professor Lars Vedel Kessing
Danish registries represent a unique opportunity to explore enormous data amounts with respect to health data, e.g., registries of prescriptions combined with morbidity data and other patient record data with phenotypic information. Within this WP, we will focus on both patient groups' drug intake to identify comorbidity, potential side effects, and drug response. We will also use Danish population-based registries to validate the outcome of various pharmacological interventions.
WP2. Electronic Patient Data: Text mining and Machine Learning
WP-leaders: Professor Søren Brunak and Associate Professor Desmond Elliott
The aim of WP2 is to use text mining methods to extract detailed, phenotypic features from free text in Electronic Patient Records (EPRs). We want to use EPRs from two sources; the Capital region and region Zealand from 2009-2018 and from the specialized national epilepsy hospital, Filadelfia.
WP3. Deep Phenotyping Data from Established Research Cohorts
WP-leaders: Professors Vibe G. Frøkjær and Kamilla Miskowiak
Whereas we can retrieve clinical and biochemical data from patient records, additional features can be identified from existing research databases and biobanks hosted in the Capital Region. Such inventories include the Lundbeck Foundation Center Cimbi database and data acquired by the Kamilla Miskowiak group. These databases will be exploited to identify biomarkers that are predictive of symptom resilience or vulnerability, or treatment outcome; e.g., certain genetic, epigenetic, cognitive, molecular and functional neuroimaging features. These existing cohorts are particularly important because they contain deep phenotyping data from a large number of healthy controls which serve as an important reference for our patient studies. They also uniquely enable us to conduct register-based follow-up studies to establish which features in clinically healthy individuals can predict later development of depressive episodes; information which can be extracted from the national registries.
WP4. Deep Phenotyping Data from New Research Cohorts
WP-leaders: Professors Lars Pinborg and Martin Balslev Jørgensen
To increase power to detect biomarkers and treatment response, we will establish new cohorts of patients with epilepsy and MDD that we follow longitudinally. With the experience gained from the other WP’s, such cohorts will enable replication of previous findings and empower additional research findings. Read more about the new longtudinal epilepsy and depression studies via the following links:
The BrainDrugs-Epilepsy Study - Precision medicine in the treatment of epilepsy (BrainDrugs-E)
Precision Medicine in the Treatment of First Episode Depression – establishing the BrainDrugs-Depression Cohort (BrainDrugs-D)
WP5. Mining neuroimaging data
WP-leaders: Associate professor Melanie Ganz and professor Gitte Moos Knudsen
The aim of WP5 is to implement new tools to facilitate regulated access to the large volume of neuroimaging data collected during routine clinical care and available in the institutional archives. Tools to identify, access and combine neuroimaging data with data extracted from the electronic healthcare records including the medical images will be implemented through a user-friendly portal that enables interactive analysis and exploration of such valuable image repositories. Following this, the combined data will be validated by e.g. comparing automatically extracted image features with data extracted from the electronic healthcare records. Next, we will use image analysis and pattern recognition tools on the existing data to define characteristic features of poor drug responders compared to healthy controls. Finally, the neuroimaging data obtained in WP4 will be used to test and validate algorithms that are predictive of long-term outcome and drug treatment response.
WP6. Treatment Response and Genomic Data
With this WP, we aim to assess the (additional) effect of molecular alterations in selected relevant genes as well as amalgamated scores with discrete brain functions, clinical and biochemical features and treatment response of the patients.
WP7. Implementation in the Clinic
The work in this WP is to ensure a smooth and swift implementation of the research outcomes generated by this thematic alliance into the clinic, to generate a true precision medicine approach to future patients with epilepsy and/or MDD. Since this work is naturally embedded in the other work packages, especially in WP4 and WP5, we have over the first years of BrainDrugs come to the conclusion that it no longer makes sense to have WP7 represented as a separate work package.